Research
Why study DNA Mismatch Repair?
- Mutations in human mismatch repair proteins are associated with hereditary nonpolyposis colorectal cancers and other sporadic cancers.
- Mismatch repair deficient cells resist the cytotoxic effects of chemotherapeutics, specifically methylating agents like cisplatinin.
- Aberrant processing of insertion/deletion errors causes trinucleotide repeat sequences to expand, leading to neurodegenerative diseases like Huntington’s disease and myotonic dystrophy.
DNA mismatch repair (MMR) is one of several DNA repair processes that are vital for maintaining genome stability. In DNA synthesis, a mismatch (non-Watson-Crick base pairing) occurs once in every 106-107 base pairs. The MMR system has been shown to improve the fidelity of DNA synthesis (by post-replicative methods) by 100-1000 fold. The MMR machinery is able to correct not only mismatches but also insertion-deletion loops (IDLs) which may occur during replication and recombination of DNA.
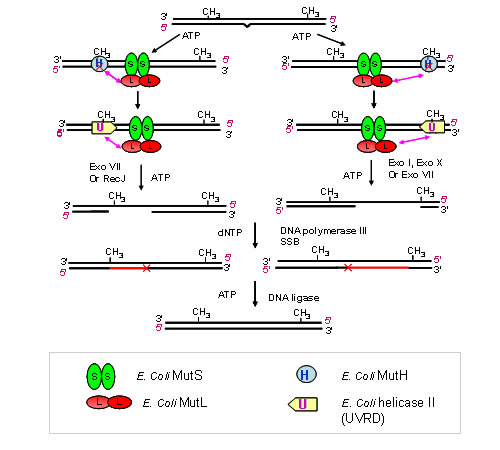
MMR is also able to correct DNA damages caused by internal or external sources. Inactivation or deterioration of this pathway has been linked to a variety of cancers, most notably Hereditary Non-Polyposis Colorectal Cancer. The process of MMR is best understood in Escherichia coli (E. coli), where it is carried out by the mismatch repair proteins MutS, MutL and MutH along side the replication machinery. Hemi-methylation of a GATC site provides discrimination between the parental and daughter strand of DNA.
What do we study?
The two initiation proteins in MMR, MutS and MutL are conserved from prokaryotes to eukaryotes. Unlike the homodimeric prokaryotic proteins, there is an array of heterodimers that function not only in mismatch repair but also in meiotic recombination. Since there is no known MutH homolog and no methylation occurs in eukaryotes, the mechanism of strand discrimination is unknown. However, recent research has reconstituted yeast MMR in vitro.
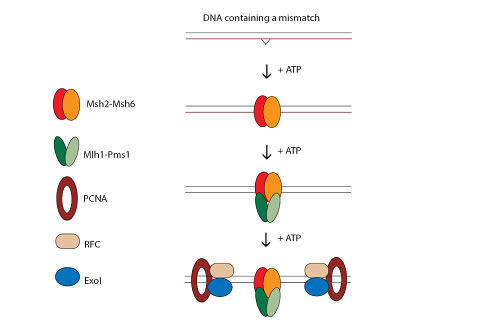
Figure: Simplified version of DNA MMR initiation in S. cerevisiae. Msh2-Msh6 binds to a mismatch in an ATP-dependant fashion. Mlh1-Pms1 is then recruited in an ATP-dependant fashion. PCNA, RFC and ExoI all take part in the excision of the DNA containing the mismatched base.
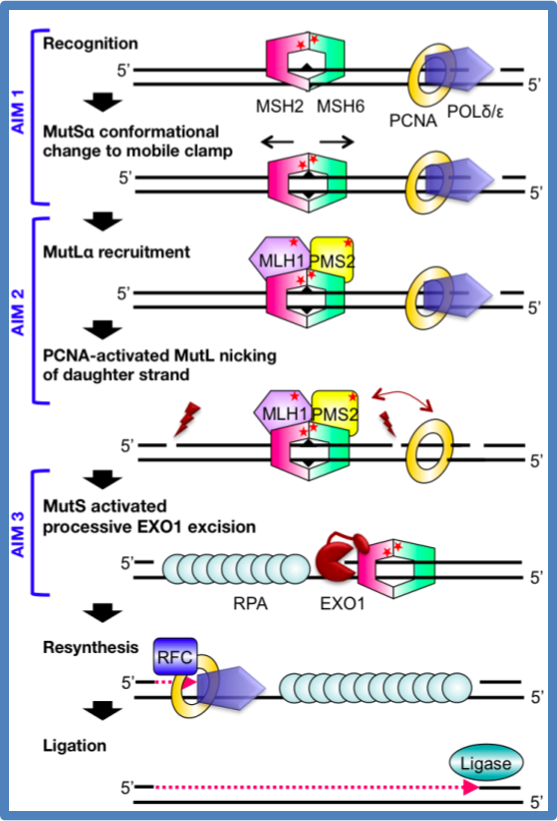
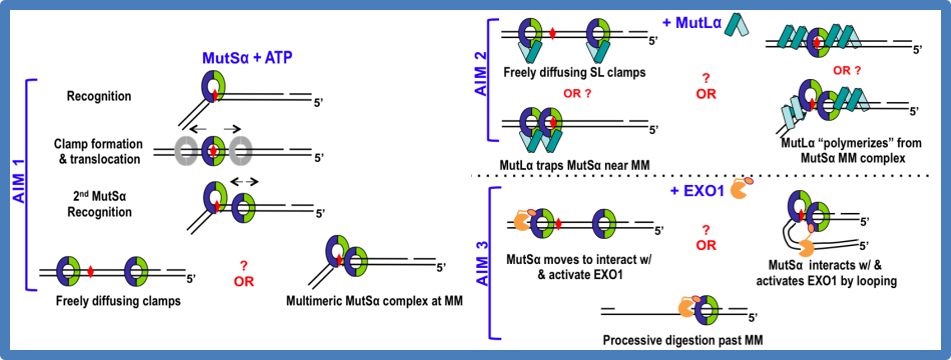
Our lab focuses on the initial steps of mismatch repair. We seek to describe the structure-function relationships that govern the protein-DNA complexes involved in error recognition, strand discrimination, and excision of the error. The following diagram lists quests (in question marks) our lab is currently seeking to understand.
Figure: Hypothesized models in MMR
We additionally are looking at other protein-protein and protein-DNA interactions related to DNA repair, including interactions of RFC and PCNA and MutS mutants with mismatched DNA.
You can find a list of current and past studies on DNA mismatch repair below, or navigate to the ‘DNA Mismatch Repair’ Topics section on the left sidebar.
-
Posts related to DNA Mismatch Repair
- Combined AFM and Fluorescence Microscopy TechniqueAFM is a powerful technique for studying protein-protein and protein-DNA interactions. However, a significant limitation to AFM and other high-resolution microscopy is the difficulty of identifying different proteins in multi-protein complexes. To resolve this, we are interested in combining the ...More
- AFM Studies of DNA Repair – OverviewAFM provides us unique capability in revealing structural and functional relationship of DNA repair, including but not limited to, the specificity of the protein binding complex, the binding affinity, the DNA bending through bend angle analysis, the conformation analysis, and how ...More
- Single Molecule FRET Studies of DNA Repair Dynamics – OverviewWe have developed single-molecule FRET methods to study conformational dynamics of DNA-protein complexes as well as to analyze large multi-protein complexes related to DNA repair. Motivation Our group focuses primarily on structure-functions studies of the DNA mismatch repair pathway. Although our group has ...More
- Combined AFM and Fluorescence Microscopy Technique
In the meanwhile, our lab collaborates with other labs in various other research topics.
- Transcription Transcription is the DNA directed synthesis of RNA, and is the first step in a series of events that leads to gene expression.
- Nucleosome Nucleosomes are DNA packaging units that are the fundamental building blocks of eukaryotic genomes.
- DNA Mismatch Repair DNA mismatch repair is a system for recognizing and repairing erroneous insertion, deletion, and mis-incorporation of bases that can arise during DNA replication and recombination, as well as repairing some forms of DNA damage.