Research: Atomic Force Microscopy, Past Studies
Topic: DNA Mismatch Repair
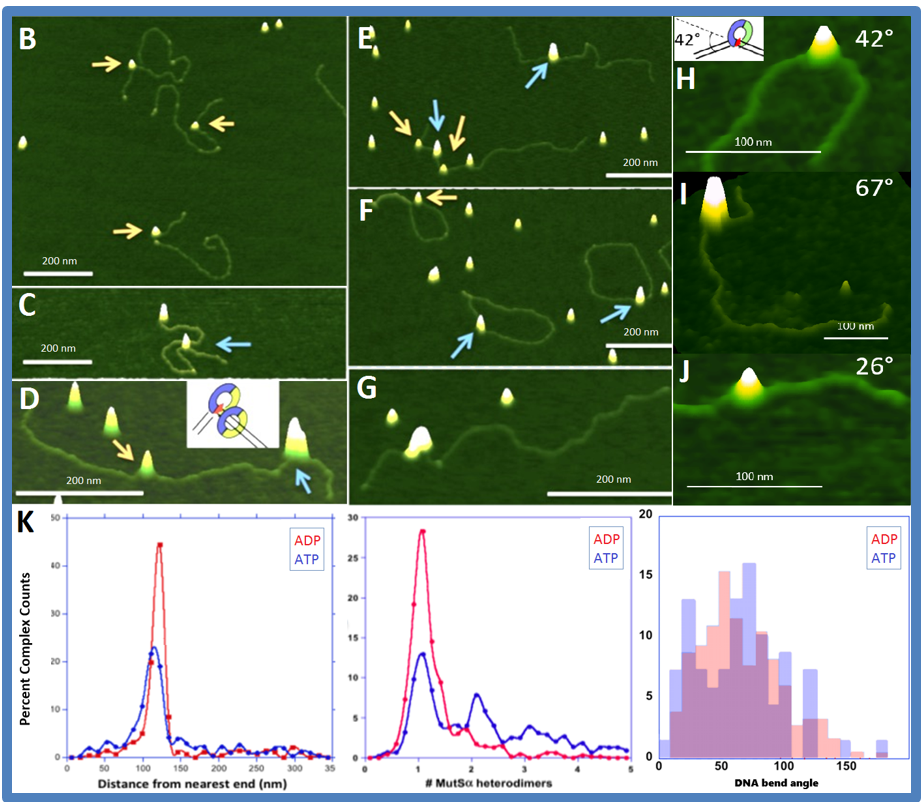
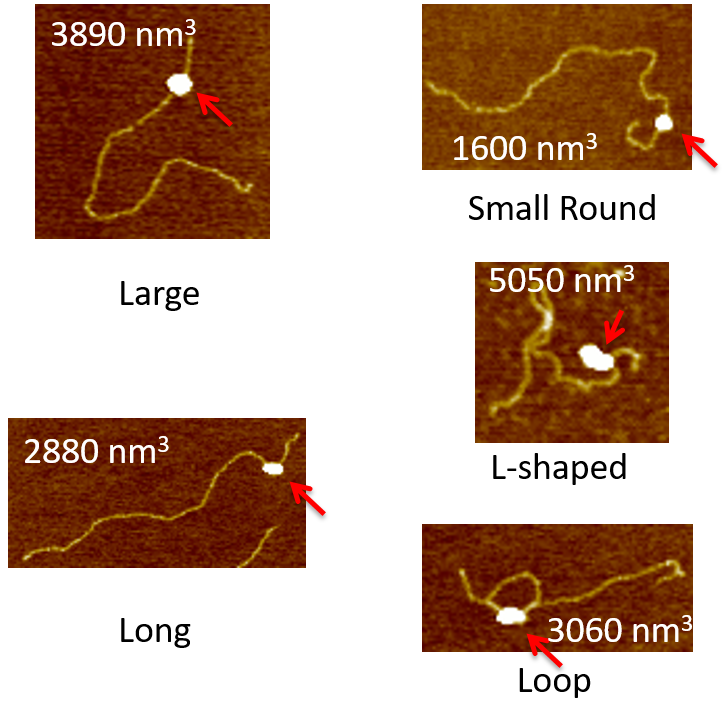
AFM provides us unique capability in revealing structural and functional relationship of DNA repair, including but not limited to, the specificity of the protein binding complex, the binding affinity, the DNA bending through bend angle analysis, the conformation analysis, and how they relate to the biological context of its functions. The following figure gives you a walk-through over what image information we are seeking and the analysis that we could get from these images.
[label type=”label-default”]
Project from Kira
[/label]
MutS
The DNA binding properties of MutS and its homologs, the initiation protein in the DNA mismatch repair pathway, is the center piece of our AFM studies. For example, we use atomic force microscopy to examine the DNA binding properties of yeast Msh2-Msh6, the initiation protein in the DNA mismatch repair pathway.
[label type=”label-default”]
Project from Susan
[/label]
These results support our model for prokaryotic mismatch repair initiation, which shows that the DNA exists in two conformations when MutS is bound to the mismatch: one in which the DNA is bent and one in which the DNA is unbent. We are now able to relate structure-function properties of mismatch repair initiation in prokaryotes with eukaryotic MutS homologs.
MutL
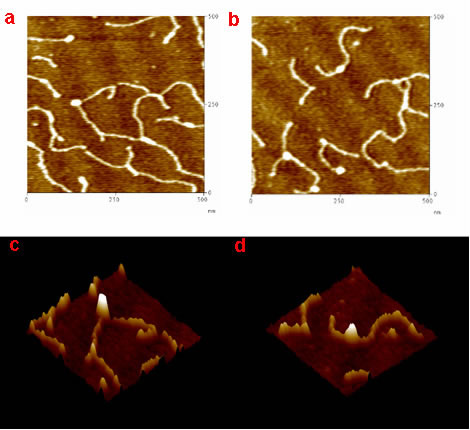
We also study the downstream mismatch repair protein MutL and its homologs. For example, we use atomic force microscopy to image Saccharomyces cerevisiae yMutLα under a variety of conditions (presence and absence of DNA and/or adenine nucleotide, varying salt concentrations) in order to understand the role yMutLα plays in mismatch repair.
[label type=”label-default”]
Project from Liz
[/label]
[label type=”label-default”]
Project from Junghoon
[/label]
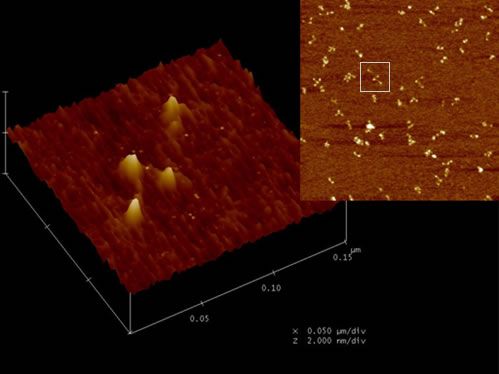
MutS-MutL
We also use AFM to study the interaction of MutS and MutL related to DNA mismatch repair initiation. MutS-MutL complexes recruit downstream proteins in the mismatch repair system to complete the repair process.
[label type=”label-default”]
Project from Kira
[/label]