Research: Kinetics, Past Studies
Topic: Transcription
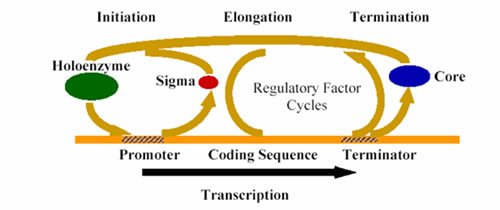
Transcription is the DNA directed synthesis of RNA, and is the first step in a series of events that leads to gene expression. In both prokaryotes and eukaryotes, an evolutionarily conserved class of enzyme known as RNA polymerase carries out the transcription reaction. The process of transcription consists of three phases: initiation, elongation, and termination, see Figure 1. In the Erie lab, we are most concerned with transcription elongation.
Throughout the process, RNA polymerase (RNAP) must carry out transcription at a reasonable rate and with a low occurrence of incorrect incorporations to maintain the necessary expression of genes within a cell. To better understand the process and regulation of transcription, it is necessary to fully characterize each step in the transcription pathway and determine the rate limiting steps for both correct and incorrect nucleotide addition. For this characterization, transient-state kinetics are used to investigate the individual steps in the transcription mechanism.
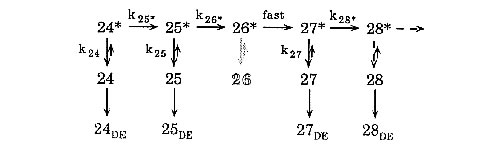
Erie et al used in incorrect incorporation, a.k.a. misincorporation, experiments to determine the initial branched mechanism of transcription elongation (Figure 2). The mechanism suggests that synthesis involves an activated (n*) and unactivated (n) enzyme complex. The transition from the unactivated to the activated state is characterized by conformational changes in the RNAP. The activated state is long-lived with synthesis occurring rapidly following NTP binding. Complexes can also decay off of the activated pathway and be trapped in the non-productive unactivated state (Erie, Hajiseyedjavadi et al 1993). Similar results have also been seen with eukaryotic RNA polymerase II (Thomas, Platas et al 1998).
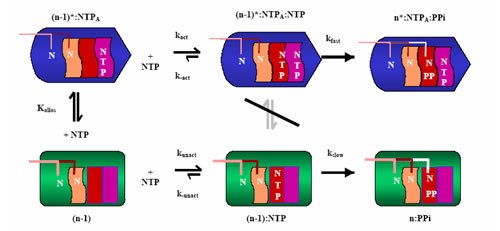
Further characterization of the kinetic mechanism using correct incorporation studies suggest that the transition from the unactivated to the activated state is achieved by nucleotide binding to a separate allosteric site on the RNA polymerase (Foster, Holmes et al 2001). Utilizing this information, an updated mechanism has been proposed (Holmes & Erie 2003).
The mechanism shown in Figure 3 suggests that the pre- and post-translocated states of the enzyme are in rapid equilibrium. Binding of an NTP shifts the equilibrium towards the post-translocated state. An NTP binding first to the allosteric site forces the enzyme to enter the activated state of transcription. The switch between the activated and unactivated state is caused by a conformational change in the enzyme induced by the NTP binding in the allosteric site. After activation, the next NTP to be incorporated enters the catalytic site, and pyrophosphate is released as synthesis occurs. However, if an NTP binds first to the catalytic site, the enzyme remains in the unactivated state (Holmes & Erie 2003).