Research: Kinetics, Past Studies
Topic: Transcription
Utilizing pre-steady state kinetics with various other biochemical techniques, the Erie lab worked toward a better understanding of the process and regulation of transcription elongation. Our past projects included:
- Correct incorporation experiments investigating the effect of incubation with downstream nucleotides as well as correct and incorrect kinetic studies using variant RNA polymerases that are specifically designed to elucidate the location of the proposed allosteric site
- Incorrect incorporation kinetics using wild type and mutant RNA polymerases that are designed to investigate changes in and around the secondary channel of the enzyme
We use pre-steady state enzyme kinetics to study incorrect incorporation by E. coli RNA polymerase during transcription elongation. This research also includes characterization of mutant E. coli RNA polymerase known to have an increased rate of incorrect incorporation.
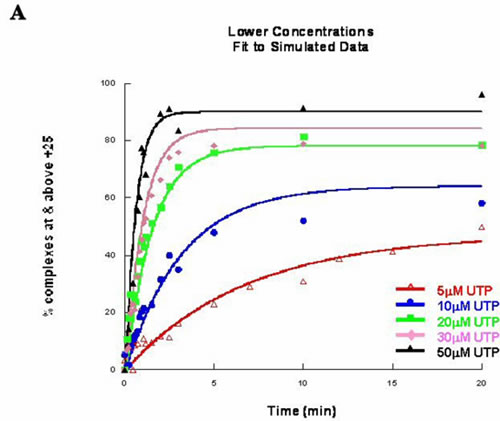
Figure: A) Concentration dependent misincorporation kinetic data for [UTP] = 5µM to 50µM fit to proposed kinetic mechanism. B) Concentration dependent misincorporation kinetic data for [UTP] = 50µM to 600µM fit to proposed kinetic mechanism.[label type=”label-default”] Project from Candi[/label]
- GreA and GreB factor-mediated transcript cleavage reactions as part of the study of the regulation mechanism for transcription
- Nucleotide binding studies to determine the number of NTP binding sites on E. coli RNA polymerase with molecular modeling and molecular dynamic simulations to visualize protein-protein and protein-DNA interactions
Single-molecule studies have shown that RNA Polymerase (RNAP) transcribes RNA with varying rates from molecule to molecule. Our current research focuses on using NTP-binding information to resolve various rates of transcription and compare those rates with those defined in the kinetic mechanisms obtained from transcription reactions in bulk solution.
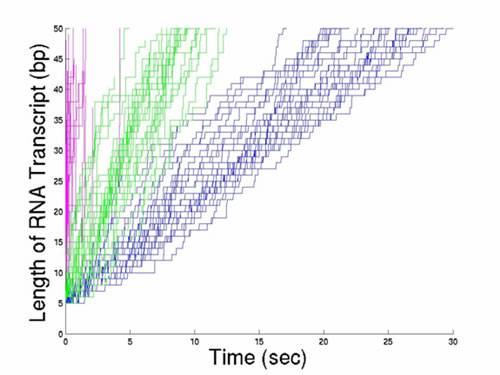
Figure: The figure above illustrates a Monte Carlo simulation of a potential mechanism for transcription elongation of single RNA transcripts at three different NTP concentrations. The magenta lines corresponds to [NTP] = 2 mM. The concentrations for the green and blue lines are 100 and 10 micromolar, respectively. These simulations suggest that the rate of transcription varies more from molecule to molecule at lower NTP concentrations than at high NTP concentrations.[label type=”label-default”] Project from Cherie[/label]
- Development of novel separation of free NTP’s from large biological molecules such as RNAP
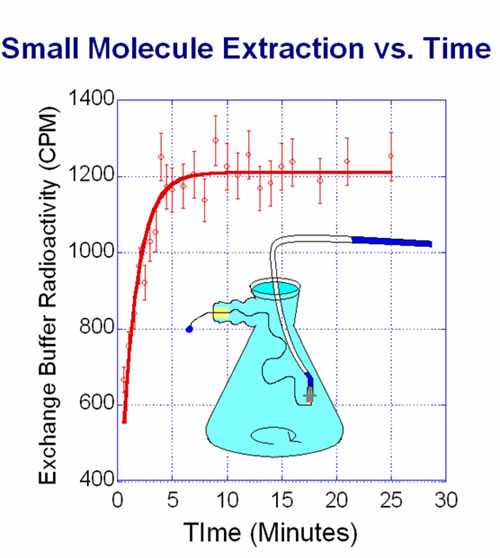
We are developing techniques to separate free NTPs from large biological complexes with high efficiency. Several avenues are being pursued, including the use of dialysis fibers and electromigration through size discriminating media. There are several advantages to removing background molecules from small molecule/protein complexes completely and quickly.
Since binding constants are usually determined via some type of identifying marker on the small molecule bound to a complex, reducing the number of background small molecules will reduce the signal to noise of the experiment. Higher purity of these complexes will allow for the determination of smaller binding constants (like that of many ATP dependant systems) and of course, fast “sample clean-up” will open a window for kinetic studies on faster time scales.
[label type=”label-default”] Project from Bob[/label]