Research: Single Molecule FRET
Topic: DNA Mismatch Repair
We have developed single-molecule FRET methods to study conformational dynamics of DNA-protein complexes as well as to analyze large multi-protein complexes related to DNA repair.
Motivation
Our group focuses primarily on structure-functions studies of the DNA mismatch repair pathway. Although our group has done tons of work looking at single DNA-protein complexes and protein-protein complexes using AFM. There are some limitations to AFM, including the inability to directly measure conformational dynamics or to decipher large multi-protein complexes.
AFM studies and crystallographic data have been used to study interactions of mismatch repair factor MutS with mismatched DNA. We want to better understand how MutS may recognize a single base mismatch when hundreds upon thousands of correctly-paired bases are present. The crystal structure of MutS bound to mismatched DNA showed that MutS induced a kink in the DNA at the mismatch, which may play a role in how MutS recognizes mismatches over homoduplex DNA. This structure does not explain why some mismatches are repaired more efficiently than others.
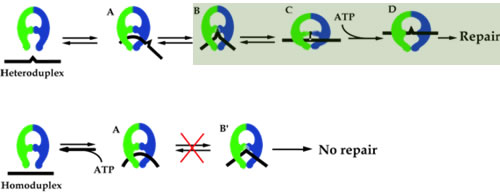
Our group sought-out to relate DNA structure with repair efficiency. AFM results showed that when MutS was bound at a DNA mismatch, two DNA conformations ensued: bent and unbent. Because only mismatched DNA displayed the unbent conformation, we developed a model for mismatch repair initiation that suggested that MutS first bent the DNA at the mismatch, and this bent conformation drove the formation of the unbent conformation, which was the conformation that went on to be repaired in the cell. Our model, shown in Figure 1, suggests that it is totally essential for the unbent complex to form for the DNA to be repaired.
Because AFM does not give us dynamic changes in the DNA conformation, we have to seek other methods to understand the dynamics in DNA repair.
smFRET
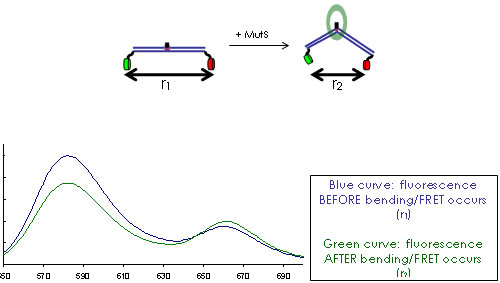
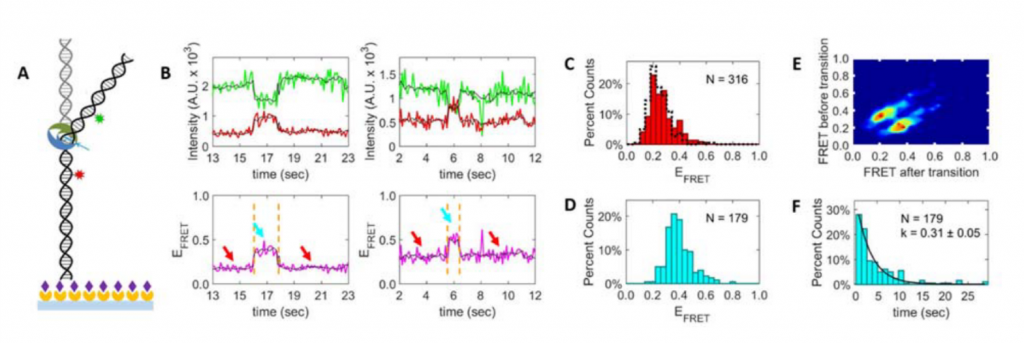
FRET is the perfect tool to understand the dynamic equilibrium between bent and unbent DNA conformations (Figure 2,3) in the presence of MutS and how this equilibrium is affected by other repair proteins and cofactors in the cell. We use TIRF microscopy to measure single-molecule FRET of mismatched DNA molecules in the presence and absence of MutS. We want to learn more about the DNA conformational dynamics, bending and unbending, that may be essential to proper mismatch repair. This project is in collaboration with Keith Weninger at NCSU Department of Physics. With FRET, we are also studying the intra-molecular dynamics of DNA repair proteins, in particular how their conformational states (e.g. open or closed) correlate with DNA binding and bending events on and off the mismatched site.
[label type=”label-default”]
Image credit: Lauryn
[/label]
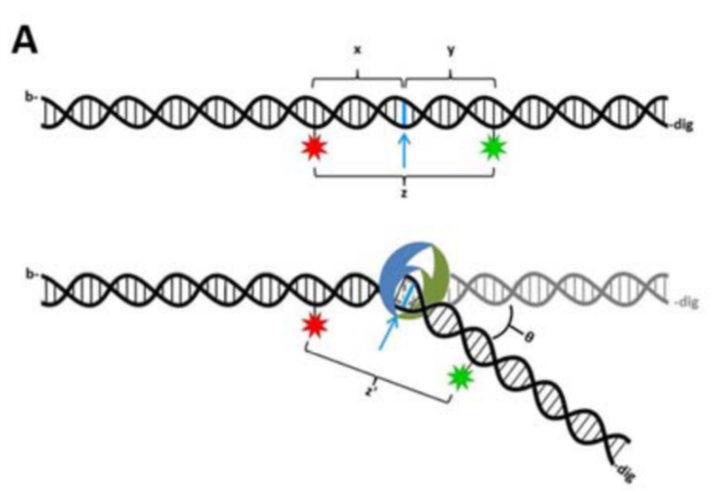
Figure 3. MutS bends the DNA at the mismatch, which brings the donors and acceptors (both engineered on the DNA) closer and activates FRET.
[label type=”label-default”]
Image credit: Jake
[/label]
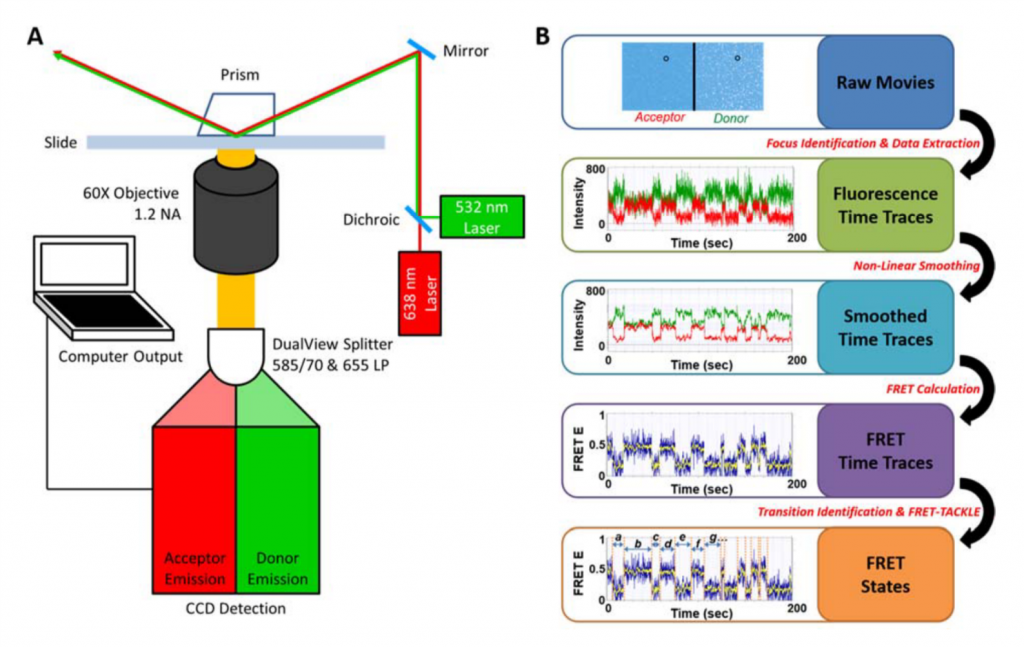
Our students developed in house algorithms and a novel workflow for our data analysis that allows for high throughput analysis (Figure 4).
[label type=”label-default”]
Image credit: Jake
[/label]
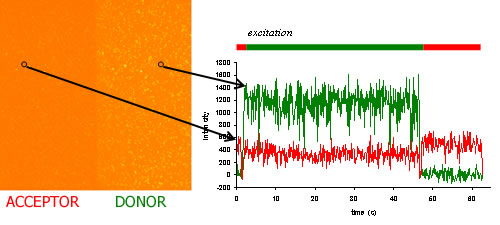
With smFRET, we can categorize conformational states based on their FRET values, and transitions of states based on their dynamics. Example analyses are shown below.
[label type=”label-default”]
Image credit: Jake
[/label]
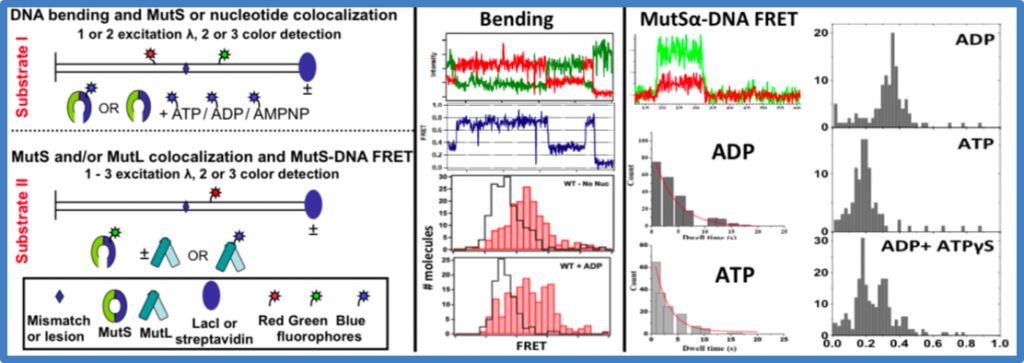
Figure 6. Data analysis – capturing MutS-DNA interactions with FRET
(left). Substrates for bending (I) and MutSa-DNA FRET (II) experiments. Time traces of donor (green) and acceptor (red) fluorescence intensities (top) and FRET efficiencies (center) for a MutS-GT complex. FRET distributions for MutSa binding and bending a GT (using Substrate I) in the absence and presence of ADP (bottom). The skyline plots are free DNA.
(middle). Fluorescence intensities for ReAsH-MutSa binding to Cy5-GT using Substrate II (top). Dwell time distribution of ReAsH-MutSa-GT complex in the presence of ADP and ATP. Calculated lifetimes for ADP and ATP are 1.8 and 4.3 s, respectively.
(right). Distributions of FRET efficiencies for Cy3-SplAsH-MutS bound to Cy5-GT in ADP (top), ATP (middle), and ADP+ATPgS (bottom) using Substrate II.
We have also used AFM to study how MutS and MutL (the first two repair factors and the most highly conserved of all the repair factors) interact at a mismatch to invoke repair. We found that multi-MutL-MutS complexes form large tracks along the DNA. We are not able to differentiate between MutS and MutL molecules in these large complexes, so we would like to combine AFM with fluorescence to add a new dimension of information to these images. We are interested in combining AFM with FRET to get a topographical image of complex multi-protein systems as well as a fluorescence image to tell us which protein is which, and how many are involved in the complex.